More of the DNA in the human genome is transcribed into RNAs than scientists can adequately account for. Transcription of around 20,000 known protein-coding genes covers about 40% of the genome, but at least 75% of the genome is transcribed reproducibly at a detectable level1,2. A decades-old debate in genomics has failed to resolve how much of the extra RNA transcribed — including thousands of long non-coding RNA sequences — is functional, and how much is ‘noise’3,4. Central to the disagreement is a lack of clarity about the nature of this transcriptional noise5. In 2013, I suggested a ‘Random Genome Project’ to establish a baseline expectation for the biochemical activity of genomic DNA in the absence of any evolutionary selection for biological functions6. Fuelled by rapid advances in synthetic genomics, two studies, one in Nature7 and one in Nature Structural and Molecular Biology8, describe versions of this experiment in yeast (Saccharomyces cerevisiae) and in mammalian cells.
Read the paper: Synthetic reversed sequences reveal default genomic states
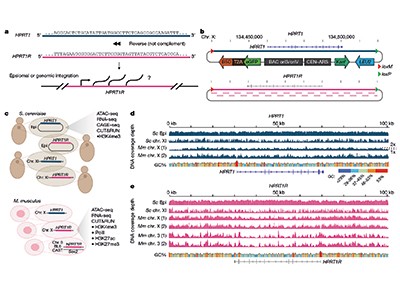
A Random Genome Project would involve synthesizing a large swathe of DNA with a statistically random sequence and running the usual high-throughput genomics assays on it. Such experiments were not technically feasible at the time they were first proposed, but are possible today. Using large-scale genome synthesis methods that researchers in their laboratory helped to pioneer, Camellato et al.7 constructed a piece of synthetic DNA that was 101 kilobase pairs in length, made of the reversed, not complementary, sequence of the human HPRT1 gene. They integrated this reversed-sequence construct into the genomes of yeast and into two sites in the genomes of mouse embryonic stem cells.
To assess transcriptional activity, the authors measured expression of RNA and the accessibility of DNA to transcriptional machinery. They also looked at two marks that signify the addition of methyl groups (methylation) to proteins called histones, around which DNA is packaged as chromatin. These marks, referred to as H3K4me3 and H3K27me3, are associated with transcriptionally active and repressed chromatin states, respectively. The bottom line is that the reversed sequence is extensively transcriptionally active in yeast — but nearly silent in the mouse cells (Fig. 1).
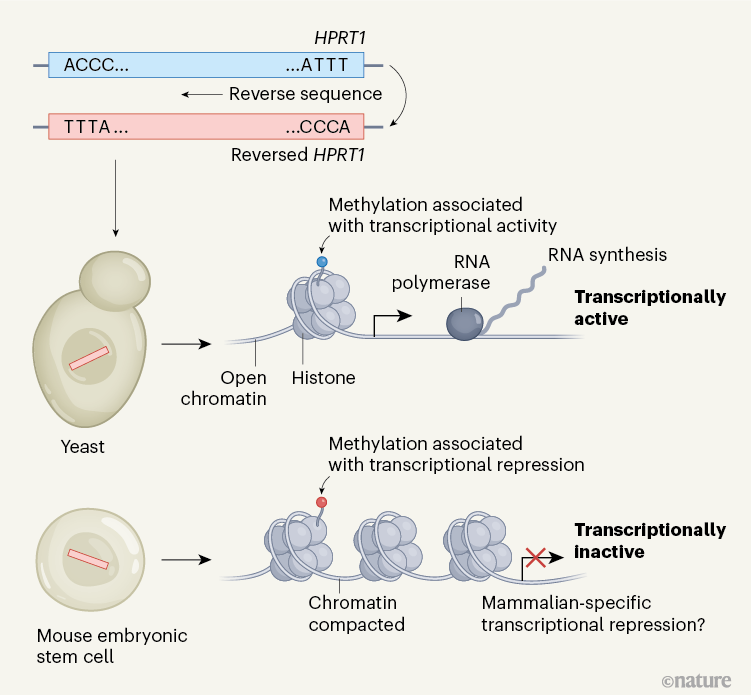
Figure 1 | Transcriptional activity of random DNA sequences integrated into yeast and mouse genomes. In many organisms, more of the genome is transcriptionally active than would be expected if only known genes were transcribed. To find out whether the extra RNA transcribed is just ‘noise’ or not, Camellato et al.7 and Luthra et al.8 conducted versions of a Random Genome Project to examine the baseline transcriptional activity of large pieces of DNA with effectively random sequences. Camellato et al. reversed the sequence of the human HPRT1 gene and integrated it into the genomes of yeast cells or mouse embryonic stem cells. They then measured signs of transcriptional activity: RNA synthesis and recruitment of the protein (RNA polymerase) that mediates it; the accessibility (open or compacted) of DNA in complex with histone proteins, which together form chromatin; and the methylation state of histones. Both studies found that random DNA was transcriptionally active in yeast, but Camellato et al. found that it was almost completely transcriptionally inactive in mouse cells. This suggests that the default state of the mammalian genome is more ‘off’ than is the case in yeast, possibly because mammals have evolved more mechanisms to repress spurious transcription.
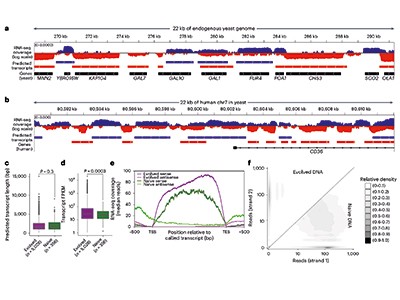
In a related set of experiments, Luthra et al.8 introduced two large pieces of human DNA (760 kb and 811 kb) as yeast artificial chromosomes, on the assumption that humans and yeast are so evolutionarily diverged from each other that human DNA would effectively seem like a random sequence to the yeast transcriptional machinery. As in the study by Camellato et al., Luthra et al. find that this ‘random’ DNA shows extensive transcriptional activity in yeast. To address what happens to random DNA in mammalian cells, they used a state-of-the-art computational deep-learning method for inferring mammalian transcriptional features to predict that synthetic random sequences should also be transcriptionally active in mammalian cells. Luthra and colleagues’ computational predictions highlight that the surprise is not that the random sequence is transcriptionally active in yeast, but that Camellato et al. find that random sequences are not very active in mammalian cells.
Read the paper: Regulatory activity is the default DNA state in eukaryotes
Other studies published in the past few years have seen broadly the same result in yeast using different DNAs that are random, non-biological or not native to yeast (exogenous). The DNAs in these studies included an 18-kb synthetic, uniformly random sequence9, a 254-kb synthetic DNA that encodes a digital image file as an example of using DNA for data storage10, and exogenous pieces of DNA such as the whole genomes of the bacteria Mycoplasma pneumoniae (around 800 kb) and Mycoplasma mycoides (around 1,200 kb)11. For all of these sequences, discrete RNA products and signatures of active chromatin are observed in yeast.
Why would mammalian and yeast cells be so different in what they do with the same random DNA? The core transcriptional machinery of yeast and mammals is generally similar. The explanation might instead lie in genome surveillance systems that suppress expression of DNA of foreign origin that has been integrated into the mammalian genome throughout evolution, such as transposons and endogenous viruses. It could also lie in the RNA quality-control systems that suppress spurious RNA transcripts. Compared with yeast, maybe mammalian systems have extra or stronger noise-suppression systems to defend their larger genomes against a larger load of genomic parasites.
For example, Camellato et al. observed that the reversed synthetic DNA is marked by H3K27me3 in both integration sites in mouse cells, indicative of transcriptional repression by the Polycomb protein complex — a prime example of a repression system found in mammals but not in yeast. Polycomb recruitment in mammals correlates with the number of sites in which cytosine and guanine bases are found next to each other (the CpG dinucleotide content), which is strongly and distinctively depleted in evolved mammalian genome sequences. But Polycomb-mediated repression turns out not to be the explanation here. Camellato et al. tested this possibility by synthesizing and inserting a different reversed sequence from which every CpG dinucleotide had been removed. Bafflingly, although the reversed DNA without CpG no longer showed H3K27me3 enrichment, it remained transcriptionally nearly silent.
Expanded ENCODE delivers invaluable genomic encyclopedia
Is there another system that could be suppressing expression of RNA from the random sequence in mammalian cells? One good candidate might be the HUSH (human silencing hub) protein complex, which is found in vertebrates but not in yeast12. The HUSH complex transcriptionally silences foreign DNA that expresses RNA transcripts without introns (intervening sequences that are removed from the transcript by a process called splicing) as a general innate defence system against RNA-based foreign genetic elements called retrotransposons. The mechanism of HUSH-mediated repression is still not fully understood, but it usually correlates with H3K9 methylation of the repressed foreign DNA. The repressive H3K9 histone mark was not assayed by Camellato et al., but it would be interesting to do this in the future.
What about the big question — do the results of Camellato and colleagues mean that the default state of a mammalian genome is ‘off’, and therefore that those thousands of observed mammalian long non-coding RNAs should be considered likely to be functional? Unfortunately, the jury remains out on this. The synthetic DNA was ‘only’ 101 kb long, which is not enough to address the question definitively. Long non-coding RNA genes, by most accounts, occur at a density of around one per 50–100 kb in the human genome7. This means that even if the majority of human long non-coding RNAs arise from transcriptional noise, Camellato et al. might easily not have observed any random long non-coding RNA genes in their 101-kb random sample.
Furthermore, Camellato and colleagues’ analyses focus on fairly abundant transcripts rather than delving down into low-level transcription, which is worth noting because most long non-coding RNA genes are detected at steady-state levels that are about 100-fold lower than those of typical messenger RNAs4. Finally, the answers to questions about the cell-type specificity of noise also await experiments in more cell types than mouse embryonic stem cells. We’re going to need a bigger Random Genome Project.
Competing Interests
Table of Contents
The author declares no competing interests.
