Since the 1950s, scientists have had a pretty good idea of how muscles work. The protein at the centre of the action is myosin, a molecular motor that ratchets itself along rope-like strands of actin proteins — grasping, pulling, releasing and grasping again — to make muscle cells contract.
The basics were first explained in a pair of landmark papers in Nature1,2, and they have been confirmed and elaborated on by detailed molecular maps of myosin and its partners. Researchers think that myosin generates force by cocking back the long lever-like arm that is attached to the motor portion of the protein.
The only hitch is that scientists had never seen this fleeting pre-stroke state — until now.
In a preprint published in January3, researchers used a cutting-edge structural biology technique to record this moment, which lasts just milliseconds in living cells.
‘The entire protein universe’: AI predicts shape of nearly every known protein
“It’s one of the things in the textbook you sort of gloss over,” says Stephen Muench, a structural biologist at the University of Leeds, UK, who co-led the study. “These are experiments that people wanted to do 40 years ago, but they just never had the technology.”
That technology — called time-resolved cryo-electron microscopy (cryo-EM) — now has structural biologists thinking like cinematographers, turning still snapshots of life’s molecular machinery into motion pictures that reveal how it works.
Muench and his colleagues’ myosin movie isn’t feature-length; it consists of just two frames showing different stages of the molecular motion. Yet it confirmed a decades-old theory and settled debates over the order of the steps in myosin’s choreography. Other researchers are focusing their new-found director’s eye on understanding cell-signalling systems, including those underlying opioid overdoses, the gene-editing juggernaut CRISPR–Cas9 and other molecular machines that have been mostly studied with highly detailed, yet static structural maps.
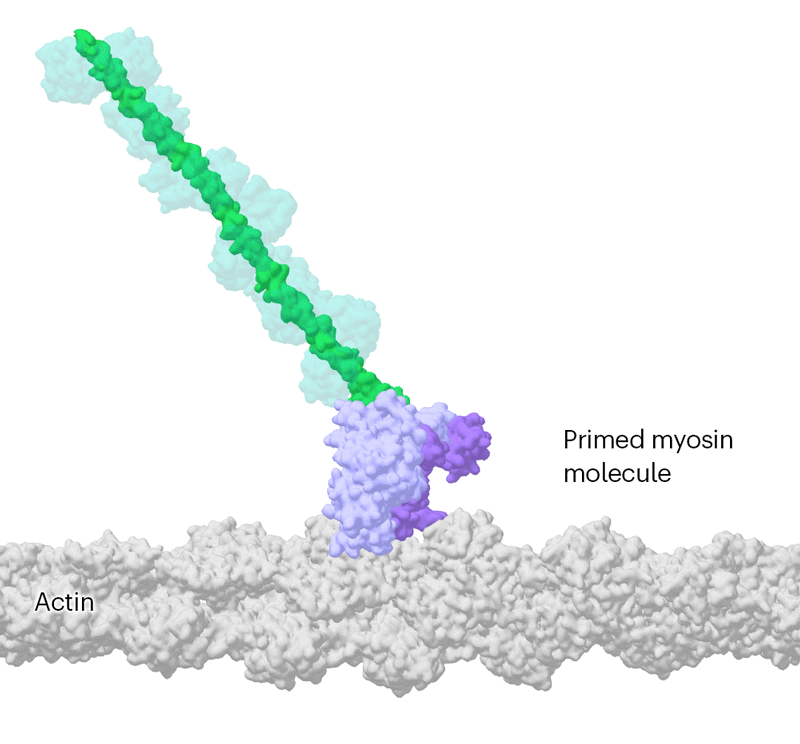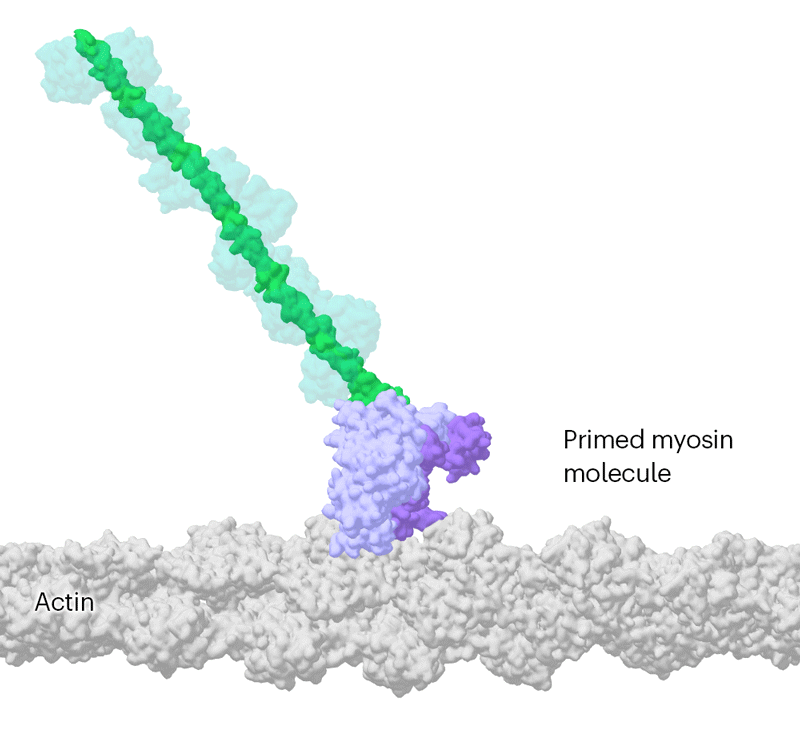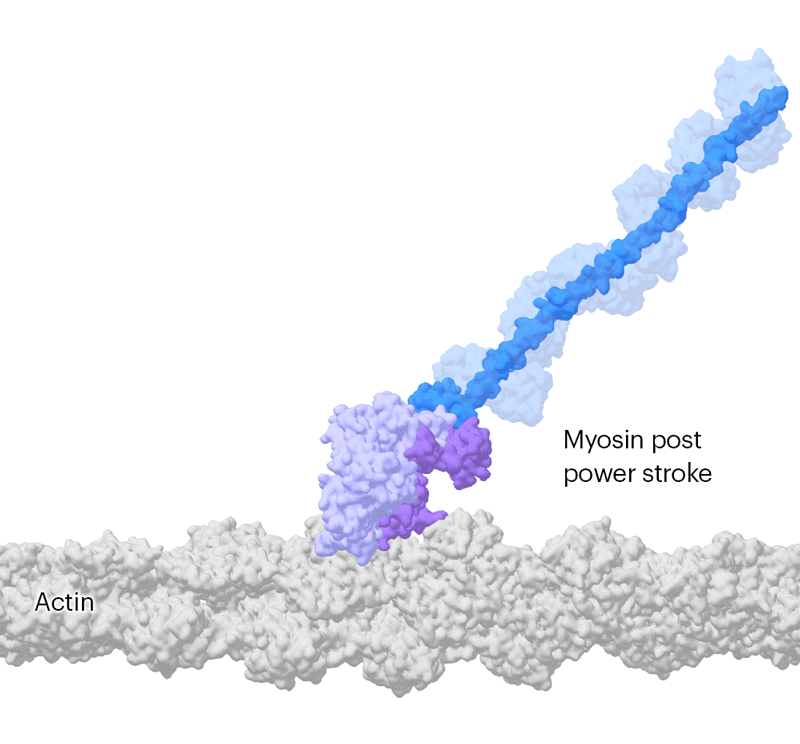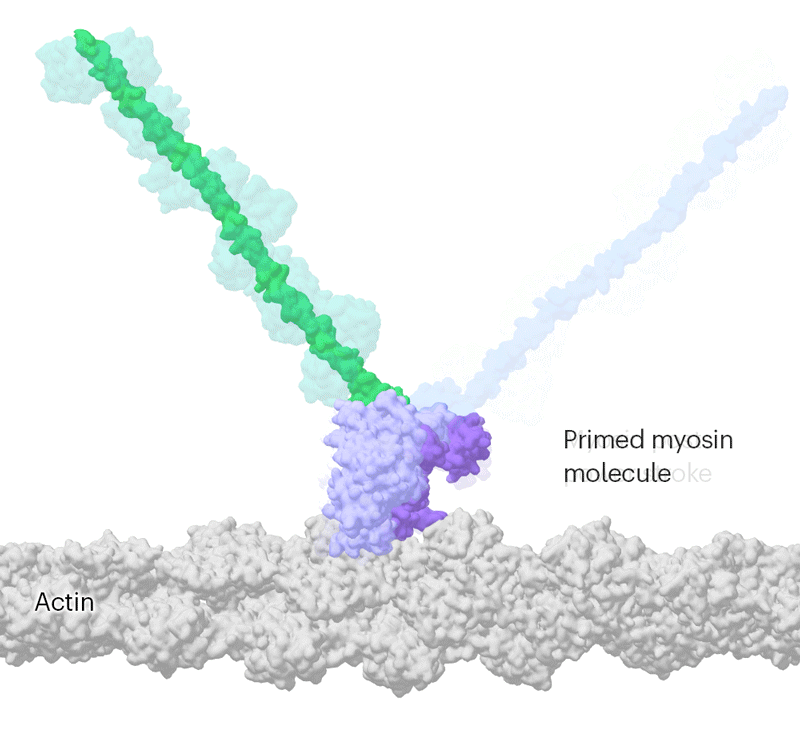
Researchers have been able to capture images of individual myosin proteins as they pull on an actin filament during muscle contraction, confirming key details of the motion. First, myosin becomes cocked or primed, then it attaches to actin and its lever arm swings in a power stroke that slides the filament by about 34 nanometres.Credit: Sean McMillan
“The big picture is to move away, as much as possible, from this single, static snapshot,” says Georgios Skiniotis, a structural biologist at Stanford University in California, whose team used the technique to record the activation of a type of cell-signalling molecule called a G-protein-coupled receptor (GPCR)4. “I want the movie.”
Freeze frame
Table of Contents
To underscore the power of cryo-EM, Skiniotis and others like to draw a comparison with one of the first motion pictures ever made. In the 1870s, photographer Eadweard Muybridge used high-speed photography technology, which was cutting edge at the time, to capture a series of still images of a galloping horse. They showed, for the first time, that all four of the animal’s hooves leave the ground at once — something that the human eye could not distinguish.
Similar insights, Skiniotis says, will come from applying the same idea to protein structures. “I want to get a dynamic picture.”
The ability to map proteins and other biomolecules down to the location of individual atoms has transformed biology, underpinning advances in gene editing, drug discovery and revolutionary artificial-intelligence tools such as AlphaFold, which can predict protein structures. But the mostly static images delivered by X-ray crystallography and cryo-EM, the two technologies responsible for the lion’s share of determined protein structures, belie the dynamic nature of life’s molecules.
“Biomolecules are not made up of rocks,” says Sonya Hanson, a computational biophysicist at the Flatiron Institute in New York City. They exist in water and are constantly in motion. “They’re more like jelly,” adds Muench.
The secret lives of cells — as never seen before
Biologists often say that “structure determines function”, but that’s not quite right, says Ulrich Lorenz, a molecular physicist at the Swiss Federal Institute of Technology in Lausanne (EPFL). The protein poses captured by most structural studies are energetically stable ‘equilibrium’ states that provide limited clues to the short-lived, unstable confirmations that are key to chemical reactions and other functions performed by molecular machines. “Structure allows you to infer function, but only incompletely and imperfectly, and you’re missing all of the details,” says Lorenz.
Cryo-EM is a great way to get at the details, but capturing these fleeting states requires careful preparation. Protein samples are pipetted onto a grid and then flash frozen with liquid ethane. They are then imaged using powerful electron beams that record snapshots of individual molecules (sophisticated software classifies and morphs these pictures into structural maps). The samples swim in water before being frozen, so any chemical reaction that can happen in a test tube can, in theory, be frozen in place on a cryo-EM grid — if researchers can catch it quickly enough.
That’s one of the first big challenges says Joachim Frank, a structural biologist at Columbia University in New York City who shared the 2017 Nobel Prize in Chemistry for his work on cryo-EM. “Even for very dexterous people, it takes a few seconds.” In that time, any chemical reactions — and the intermediate structures that mediate the reactions — might be long gone before freezing. “This is the gap we want to fill,” says Frank.
Caught in translation
Frank’s team has attempted to solve this problem using a microfluidic chip. The device quickly mixes two protein solutions, allows them to react for a specified time period and then delivers reaction droplets onto a cryo-EM grid that is instantly frozen.
This year, Frank’s team used their device to study a bacterial enzyme that rescues ribosomes, the cell’s protein-making factories, if they stall in response to antibiotics or other stresses. The enzyme, called HflX, helps to recycle stuck ribosomes by popping their two subunits apart.
Frank’s team captured three images of HflX bound to the ribosome, over a span of 140 milliseconds, which show how it splits the ribosome like someone carefully removing the shell from an oyster. The enzyme breaks a dozen or so molecular bridges that hold a ribosome’s two subunits together, one by one, until just two are left and the ribosome pops open5. “The most surprising thing to me is that it’s a very orderly process,” Frank says. “You would think the ribosome is being split and that’s it.”
Muench and his colleagues, including Charlie Scarff, a structural biologist at the University of Leeds, and Howard White, a kineticist at Eastern Virginia Medical School in Norfolk, Virginia also used a microfluidic chip to make their myosin movie by quickly mixing myosin and actin3.
‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures
But the molecular motor is so fast that, to slow things down ever further, they used a mutated version of myosin that operates about ten times slower than normal. This allowed the team to determine two structures, 110 milliseconds apart, that showed the swing of myosin’s lever-like arm. The structures also showed that a by-product of the chemical reaction that powers the motor — the breakdown of a cellular fuel called ATP — exits the protein’s active site before the lever swings and not after. “That is ending decades of conjecture,” says Scarff.
With this new model in mind, Scarff, whose specialty is myosin, and Muench are planning to use time-resolved cryo-EM to study how myosin dynamics are affected by certain drugs and mutations that are known to cause heart disease.
Microfluidic chips aren’t the only way researchers are putting time stamps on protein structures. A team led by Bridget Carragher, a structural biologist and the technical director at the Chan Zuckerberg Imaging Institute in Redwood City, California, developed a ‘spray and mix’ approach that involves shooting tiny volumes of reacting samples onto a grid before flash-freezing them6.
In another set-up — developed by structural physiologist Edward Twomey at Johns Hopkins University School of Medicine in Baltimore, Maryland, and his team — a flash of light triggers light-sensitive chemical reactions, which are stopped by flash-freezing7. Lorenz’s kit, meanwhile, takes already frozen samples and uses laser pulses to reanimate them for a few microseconds before they refreeze, all under the gaze of an electron microscope8.
‘Limitations everywhere’
The different approaches have their pros and cons. Carragher’s spray and mix approach uses minute sample volumes, which should be easy to obtain for most proteins; Twomey says his ‘open-source’ light-triggered device is relatively inexpensive and can be built for a few thousand dollars; and Lorenz says his laser-pulse system has the potential to record many more fleeting events than other time-resolved cryo-EM technologies — down to a tenth of a microsecond.
Revolutionary cryo-EM is taking over structural biology
But these techniques are not yet ready to be rolled out. Currently, there are no commercial suppliers of time-resolved cryo-EM technology, limiting its reach, says Rouslan Efremov, a structural biologist at the VIB-VUB Center for Structural Biology in Brussels. “All these things are fussy and hard to control and they haven’t really caught on,” adds Carragher.
Holger Stark, a structural biologist at the Max Planck Institute for Multidisciplinary Sciences in Göttingen, Germany, says that current forms of time-resolved cryo-EM might be useful for some molecular machines that operate on the basis of large-scale movements — for example, the ribosome. However, the technology is not ready for use on just any biological system. “You have to cherry pick your subject,” he says. “We have limitations everywhere.”
Despite the shortcomings, there are plenty of interesting questions for researchers to start addressing now using these techniques. Twomey is using time-resolved cryo-EM to study Cas9, the DNA-cutting enzyme behind CRISPR gene editing, and says the insights could help to make more efficient gene-editing systems.
Lorenz used his laser-melting method to show how a plant virus swells up after it infects a cell to release its genetic material7 (see ‘Viral blow-up’). He is now studying other viral entry molecules such as HIV’s envelope protein. “We have these static structures, but we don’t know how the system makes it from one state to the other, and how the machinery works,” he says.
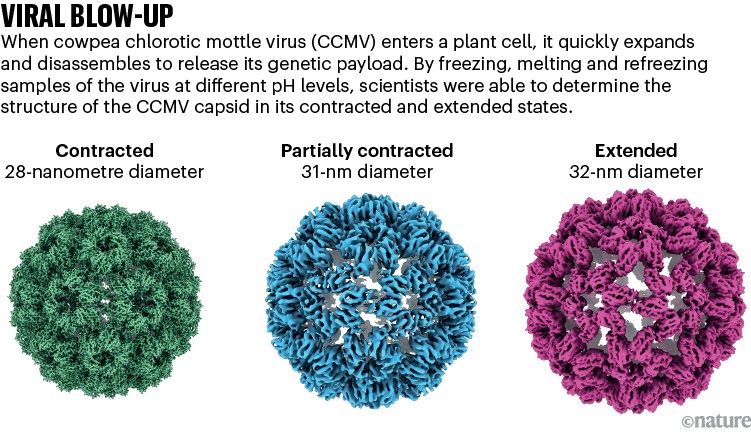
Source: Ref.8
Skiniotis’s team is investigating GPCRs, including one called the β-adrenergic receptor, which has been implicated in asthma. Their work4 shows how activating the receptor triggers it to shed its partner G-protein, a key step in propagating signals in cells.
The researchers are now studying the same process in a GPCR called the µ-opioid receptor, which is activated by morphine and fentanyl among other drugs. In preliminary unpublished results, they have found that the dynamics of the receptor help to explain why some drugs such as fentanyl are so potent in promoting G-protein activation, while others aren’t. Such insights, says Skiniotis, are glimpses of unseen biology that molecular movies promise to reveal. Just don’t forget the popcorn.
