Over the past century, the two main approaches to treating people with cancer — surgery and radiation — have focused on where in the body the tumour is. This has led to medical oncologists and other health-care providers, regulatory agencies, insurance companies, drug firms — and patients — categorizing cancers according to the organ in which the tumour originated. Yet there is a growing disconnect between classifying cancers in this way and developments in precision oncology, which uses the molecular profiling of tumour and immune cells to guide therapies.
More than ten years ago, for example, investigators in the United States showed in a clinical trial that the drug nivolumab could improve outcomes for certain individuals with cancer1. In the trial — which included people with different ‘types’ of cancer (as conventionally defined), from melanoma to kidney cancer — nivolumab shrank some people’s tumours by more than 30%, but it had little or no effect on the tumours of others.
Nivolumab targets PD1. This is a receptor of a protein called PD-L1, which helps cancer cells to escape attack from the immune system. Of the 236 trial participants whose tumours could be assessed, 49 responded positively to the treatment. The key determinant was whether their tumour cells were expressing high levels of PD-L1.

Thomas Powles: Cancer explorer
The logical next step would have been to conduct clinical trials that tested the effects of nivolumab and other PD1 inhibitors in people with metastatic tumours that strongly express PD-L1, regardless of the organ in which their cancer had originated. But because of the way cancers are classified as breast, kidney, lung and so on, researchers had to conduct clinical trials sequentially for each disease type.
For about a decade, millions of people with tumours expressing high levels of PD-L1 were not able to access relevant drugs because trials had not yet been conducted for their type of cancer when they became unwell. Those with certain breast or gynaecological cancers expressing PD-L1 had to wait 7–10 years to access PD1 inhibitors.
A similar story has played out with most of the drugs tested in clinical trials over the past decade. These include PARP inhibitors, which kill tumour cells carrying mutations in the breast cancer genes BRCA1 and BRCA2. These mutations are now known to occur in multiple tumour ‘types’ as conventionally defined (see ‘Losing lives’), not just in breast cancers.
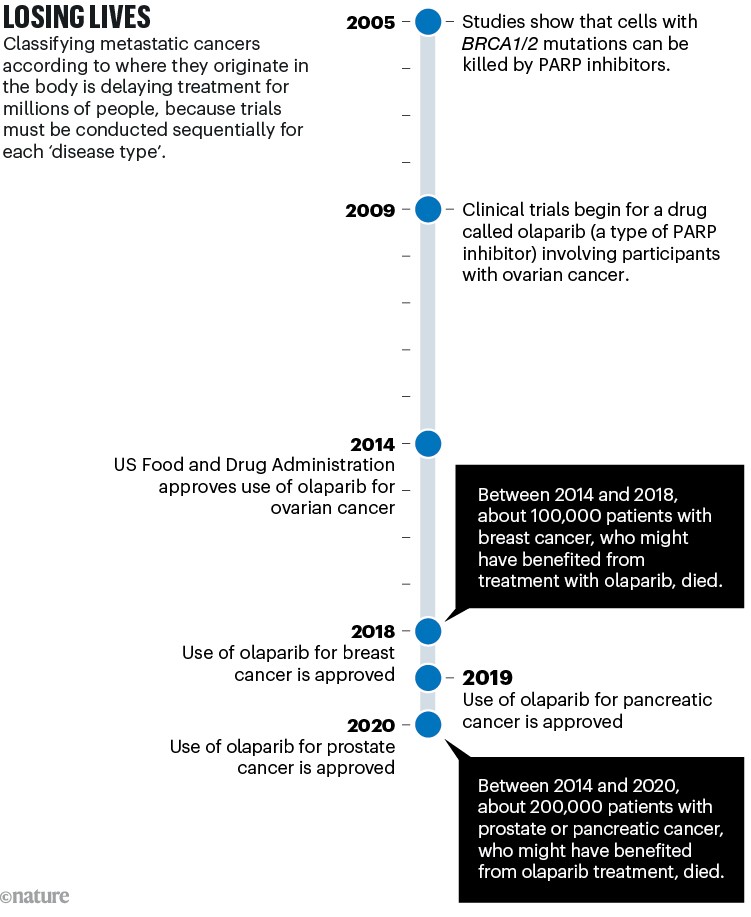
Sources: BRCA–PARP: H. E. Bryant et al. Nature 434, 913–917 (2005); H. Farmer et al. Nature 434, 917–921 (2005). Olaparib trial: P. C. Fong et al. N. Engl. J. Med. 361, 123–134 (2009). Ovarian approval: G. Kim et al. Clin. Cancer Res. 21, 4257–4261 (2015). Breast: https://go.nature.com/3STQGSU. Pancreas: https://go.nature.com/3SHNTEJ. Prostate: https://go.nature.com/4267RUJ
Metastatic cancers (those that have spread beyond the organ where they originated) account for around 67–90% of cancer deaths2,3, and are almost always treated systemically, meaning with drugs that enter the bloodstream. To improve treatments for people with metastatic cancer, the community urgently needs to shift from using organ-based classifications of cancer to using molecular-based ones. This will require radical changes in how medical oncology is structured, conducted and taught.
Habits of a century
Table of Contents
In France and some other European countries, patients are not reimbursed if they take drugs that have been tested in trials in which cancers are not defined by the organ in which they originated4. Meanwhile, most of the scientific organizations in oncology, such as the American Society of Clinical Oncology and the European Society of Medical Oncology (ESMO), organize their meetings and issue their guidelines according to ‘organ of origin’. And over the past decade, cancer centres and universities have transformed oncology into multiple organ-specific subspecialities. Hospitals have breast cancer wards, lung cancer wards and so on; medical students are offered modules in subjects such as gastroenterology and pulmonology; and preclinical research, clinical trials and treatment protocols are often tailored to organ-specific specialties.
This attachment to classifying cancer — and addressing it — on the basis of the organ in which it originated is stalling progress in multiple ways.
First, it runs counter to the scientific understanding now emerging.
The past two decades of cancer research, which have been dominated by efforts to characterize tumours at the cellular and molecular level, have shown that some of the molecular events driving their evolution are shared across different ‘types’ of cancer. Mutations in the tumour suppressor gene TP53, for example, are a feature of most types of cancer, as defined by the organ in which the cancer originated. What’s more, most cancer types can be subdivided into different molecular subgroups. Some lung cancers have mutations in the epidermal growth factor receptor (EGFR) gene, some have mutations in the MET gene, others have translocations involving the ALK gene, and so on.
Second — as already described — classifying cancer according to the organ in which it originated is making it harder for patients to obtain the drugs that could help them. In fact, when it comes to regulators approving the use of treatments, molecular-based classifications are likely to become ever more important as more drugs are developed using advanced biotechnologies.
Physicians can identify molecular targets in individual cancers and decide on treatments.Credit: Gustave Roussy
Antibody drug conjugates, for instance, are antibodies that target membrane proteins expressed by multiple types of cancer to deliver chemotherapy to tumour cells. The antibody drug conjugate trastuzumab deruxtecan has already shown promise in phase I and phase II trials in treating people whose cancers either overexpress the HER2 gene or have a mutated version of it, regardless of the organ in which their cancer originated5,6.
Last, the conventional approach to classifying cancer is hampering medical education and patient understanding.
Currently, students and practitioners have to memorize and digest an overwhelming amount of information; around 10,000 scientific articles that include the words ‘cancer’ and ‘randomized trial’ are published every year. Implementing a molecular-based classification would make it easier for students and physicians to learn. Students wouldn’t need to memorize the results of clinical trials conducted for each type because trials would be conducted across cancer types. And a knowledge of the molecular mechanisms underpinning the disease would make it easier for students to remember the outcomes of clinical trials.
The race to supercharge cancer-fighting T cells
Take, for example, a family of enzymes called PI3Ks, which are involved in cellular processes such as cell growth and proliferation. After a student is taught that these are involved in regulating glucose levels, it should be easier for them to remember that PI3K inhibitors — which are used to treat some people with breast cancer — can lead to hyperglycaemia (high sugar levels in the blood). This means that people with diabetes either should not be given these drugs, or if they are, should have their blood sugar levels closely monitored.
Molecular-based classifications could also improve people’s adherence to treatment. In our experience, the fact that any two people diagnosed with the same cancer type can be given different treatments causes confusion and misunderstanding. Most people are more familiar with body parts than with gene names. But each patient is affected by only around one to four molecular alterations, limiting the amount of new information that any one person would need to receive. And if patients are also told about the biological mechanisms driving their cancer, they will understand the rationale for treatment better. In support of this, studies from the past two decades7 have shown that telling people living with HIV why their treatment should be matched to their condition — as tracked by the count of CD4 cells (a type of white blood cell) in their blood — increases their adherence to treatment by 5%.
The way to change
Since the first trial of nivolumab in 2012, things have begun to move in a better direction, particularly when it comes to regulatory agencies approving drugs that are focused on the existence of a molecular target rather than the cancer’s organ of origin.
In 2017, the US Food and Drug Administration (FDA) approved the use of a drug called pembrolizumab to treat people who have tumour cells with a deficiency in their DNA mismatch-repair system, regardless of which organ the cancer originated in. In 2020, the agency determined that pembrolizumab could also be used to treat people whose tumour cells have high numbers of mutations relative to healthy cells and other cancer cells. And in subsequent years, it has approved the use of several other drugs to treat cancer based on the biological targets of the drugs8.
However, a much greater shift in mindset across other regulators and the cancer community at large is needed. Making this happen will require things to be done differently on at least four fronts.
Improve guidance and methodologies. Regulatory agencies, scientific societies and insurance companies need to better define what preclinical and clinical evidence is required to determine whether — when it comes to treatment — a specific molecular alteration should be prioritized over the organ in which the cancer originated.
Some scientific societies, such as ESMO, are already developing guidelines. And the FDA is working towards defining when a drug can be approved on the basis of a molecular marker, regardless of the organ in which the cancer originated9.
People with cancers that have spread beyond the organ of origin are usually treated with drugs that enter the bloodstream.Credit: Fabrizio Villa/Getty
Guidance for medical practice that has been developed in other contexts can help with this. For instance, a tool called the Magnitude of Clinical Benefit Scale allows clinicians to rank the efficacy of a drug according to various criteria. Each drug is given a score on the basis of how well people respond to the drug in clinical trials, its toxicity, its effects on the survival of trial participants and so on. Likewise, clinicians use the ESMO Scale for Clinical Actionability of Target to rank molecular alterations according to the strength of the evidence that the alteration is important when it comes to treating the person.
But a key step in developing such guidelines will be to establish the methodological and statistical tools that would enable researchers to demonstrate that a drug is working in an organ-agnostic way. How many individuals representing how many types of cancer should be included in a clinical trial investigating the effect of a drug across multiple cancer types, for instance? Or what methodology could prove that there is no difference between two tumour types in terms of responsiveness to a drug?
Restructure oncology. The problem of hospital reorganization could first be addressed by cancer centres and university hospitals, given the expertise of these institutions in molecular oncology. Such organizations could establish organ-agnostic teams that are centred around the interpretation of molecular analyses. In fact, several institutes, including the National PRecISion Medicine Cancer Center (PRISM) at the Gustave Roussy hospital in Villejuif, France, where we work, have already established teams that focus on analysing patients’ molecular profiles, regardless of cancer type.
CRISPR cancer trial success paves the way for personalized treatments
Taking this approach will be harder for small hospitals that do not have clinical departments focused on systemic therapies. But fellowships could help to transfer knowledge between institutions, and raise awareness about the benefits of prioritizing the molecular mechanisms driving cancers in treatment plans.
Rethink education. Medical students must be equipped with a comprehensive molecular understanding of carcinogenesis early in their training. This could involve asking students to come up with treatment plans focused on the underlying molecular drivers of cancer — not just to memorize the characteristics of primary tumours and the results of phase III clinical trials.
Improve access to molecular testing. The change in approach to the classification of metastatic cancer that we are calling for will not happen unless more people have access to the tests that reveal the molecular alterations in their tumour cells.
Since 2020, societies such as ESMO have recommended that all individuals with advanced lung cancer undergo multigene testing10. Yet, a study involving around 38,000 patients with this condition in the United States, who were diagnosed between 2010 and 2018, showed that only 22% had molecular test results in their medical record. This is consistent with findings from other studies, conducted both in the United States and elsewhere11,12.
Ensuring that all individuals diagnosed with metastatic cancer receive molecular testing comes down to reducing the costs of those tests. Currently, the approach costs around US$3,000 per test in the United States and around $1,000 in Europe.
But prices are falling fast: today, complete genome sequencing typically costs around $330, compared to more than $1,100 a few years ago. And some cancer centres are developing ways to perform molecular analyses themselves, so that they don’t have to rely on diagnostic companies13. Moreover, in the coming years, artificial intelligence could be used to identify genomic abnormalities from routine pathological slides at low cost14. Such innovations could make the widespread adoption of molecular testing feasible even in low- and middle-income countries.
Personalized care
In the coming years and decades, numerous layers of information could be incorporated into comprehensive characterizations of cancer that are unique to each patient.
These include the cancer’s organ of origin, which sometimes remains an important factor in deciding what treatment to try15; the number and size of tumours; and their aggressiveness, as measured by the expression levels of certain genes. Among other potentially useful information is genetic analysis of a person’s germline DNA, which can provide information about their sensitivity to certain drugs or their chances of experiencing harmful side effects; and their general health, as tracked by levels of fatigue, weight loss and so on.
Classifying cancers according to their molecular characteristics would expedite the access of millions of people to effective treatments; it is also the first step towards precision oncology and a deeper biological understanding of how cancer works.
Competing Interests
F.A.: Research Funding: AstraZeneca (Inst), Novartis (Inst), Pfizer (Inst), Eli Lilly (Inst), Roche (Inst), Daiichi (Inst). Travel, Accommodations, Expenses: Novartis, Roche, GlaxoSmithKline, AstraZeneca. Ad boards or symposium compensated to the hospital: AstraZeneca, Lilly, Novartis, Pfizer, Daiichi-Sankyo, Relay Tx and Roche. Ad board compensated to the author: Lilly. E.R.: Grants: Gilead (institutional); Travel, Accommodations, Expenses: Pfizer, Roche, Mundipharma; Honoraria: Eli Lilly, Novartis, Lilly, Seagen. A.M.: Preclinical research funding from Fondation MSD Avenir. Clinical trial drug supply from Merck Sharp & Dohme. Clinical trial drug supply and funding from Boehringer Ingelheim, Bristol Myers Squibb and Roche; personal fees, nonfinancial support from Merck Sharp & Dohme, Bristol Myers Squibb, Astra Zeneca, Roche/Genentech, Sanofi, Glaxo Smith Kline, Pfizer, Johnson and Johnson. S.M.: Personal fees for statistical advice from Amaris, for Scientific Study Committee Membership from Roche, for data and safety monitoring membership of clinical trials from IQVIA, Sensorion, Kedrion, Biophytis, Servier, Yuhan.
