My career began in the early 1990s, when computers were just a convenience in the biological sciences. Now, they’re an indispensable tool of discovery. The final year of my PhD in plant genetics, in 2000, saw the publication of the first complete plant genome, and the sudden availability of a trove of information that could be accessed only through a computer terminal. I wasn’t familiar with the term ‘bioinformatics’ at the time, but the idea that computers were essential for extracting useful information from large data sets was taking hold.
Now, I’m a professor at the California Polytechnic State University in San Luis Obispo. As a biologist, coding will be as big a part of my future — and my students’ — as extracting DNA and running gels.
I like to think I haven’t been left behind (after all, it says “Bioinformatics” next to my name on my list of research interests), so I’ve learnt to code. I’ve genuinely enjoyed it, but it hasn’t always been easy.
I started drawing cartoons as a graduate student, and most of my illustrations document biology laboratories and the people who work there. I thought they might be a good way to capture some of my experiences while learning this new skill, and that they might resonate with other experimental biologists trying to figure things out in the world of bioinformatics.
I’m often asked: what is the best way to make progress? My answer is, stand next to someone who knows what they are doing.
Imagine it: you stand a metre behind a colleague who is staring intently at their monitor. They sense you, look up from their work and warmly say, “You seem to want some help. Take a seat!” Actually, this never happens. Nevertheless, nothing increases your likelihood of making progress more than proximity to someone with more coding experience than you. Swallow your pride and (politely) interrupt them. In my experience, if you ask for help with a well-defined question and respect your colleagues’ time, they are always helpful.
Here are some other common questions, and my advice.
I got an error message. What do I do next?
Table of Contents
When I was getting started, error messages seemed to be written in another language: I could read only a few lines before my brain would shut down and dismiss the messages as ‘noise’ to be ignored.
Over time, I’ve learnt that it’s worth reading error messages, and it gets easier. It seems like obvious advice, but it did take a while for me to embrace it. Make sure, especially, to Google your errors and error messages. You’re not the first person to deal with this problem: there’s a good chance that someone else has already fixed it and posted about it.
Should I worry that my code isn’t ‘beautiful’?
You might not know why your code started working, but those lines you pasted in from your Google search seemed to do the work. If you are the only person using your code, it’s often sensible to just go with it.
It’s a nice goal to work towards code that is refined to be concise and comprehensible, will work on others’ computers and will avoid redundant computational operations. However, while you’re learning, worrying too much about beauty can hold you back. Most of my code is cobbled together.
Will I lose my lab skills?
In 2013, I did an internship with a plant-evolution research group that was entirely computational — no pipettes, bottles or plants — and it was an exciting experience. The idea of doing genuine scientific discovery with my laptop and a cup of coffee sounded idyllic at first. But after a while, running a gel started to seem very inviting. After returning to my lab, I really enjoyed finding a balance between the bench top and the laptop.
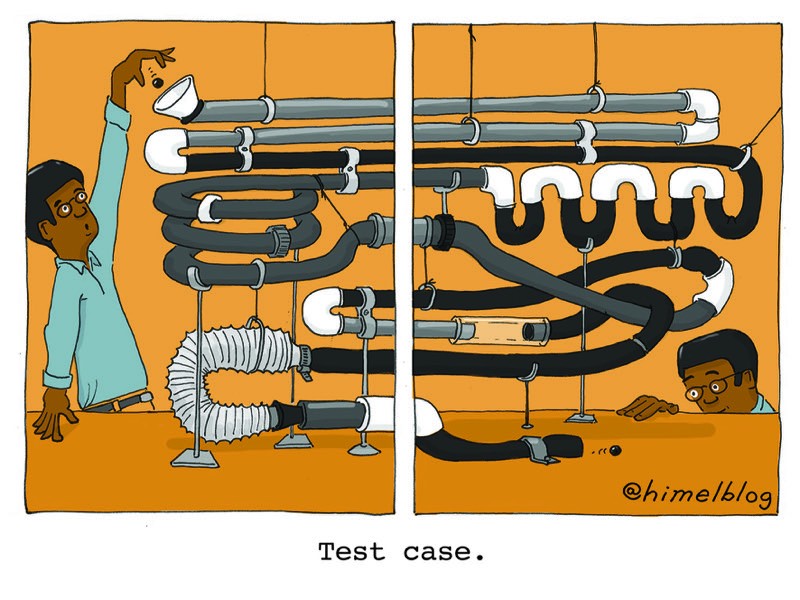
What’s the best way to test my code?
In 2015, I started teaching a bioinformatics class with a computer scientist. Biology students were paired with computer-science students; the biologists would define a question and identify the appropriate data set (often DNA sequences), then the computer scientists would write code to help solve the problem. The code would be handed back to the biologists for testing — and that’s where things often got complicated. For example, if the program was written to find the complement to a DNA strand, the biologists would paste in an entire gene sequence and then stare at the output, often missing potential problems because of the deluge of A, G, C and T.
Computer scientists and engineers are familiar with the concept of a test case: a simple set of inputs and predicted outputs that let you evaluate parts of your code quickly. For some reason, biology students learning bioinformatics have difficulty embracing this concept. Don’t start by pasting in 10 kilobytes of gene sequence. Enter “AA” — if it doesn’t give you back “TT”, you’ve got work to do.
Do I need to learn multiple programming languages?
There are many languages to choose from, each with their own strengths and applications. Should you learn them all? Or should you learn just one, and then stubbornly use it to solve every problem?
The practical solution lies between these extremes. Before joining a bioinformatics lab, I purchased books about Python and PERL, two general-purpose languages that I knew were popular among computational biologists. When I got to the lab, everyone doing the sort of work I wanted to do was using R, a language with a huge library of biology-related software ‘packages’, nuggets of pre-written code that did useful things. The Python and PERL books just sat on my desk.
The best language to learn is the one the experts around you are using. The good news is that learning one language makes it easier to pick up what you need in another (see ‘Resources’).
Is all this coding really necessary?
At some point, you’ll probably ask yourself whether time spent learning to code (troubleshooting, Googling error messages, trying and failing, and getting help from others) is really worth it. Why make that chart quickly in Excel when you could spend hours trying to make it in R?
For me, moving along the coding learning curve opened up new and interesting research, made me a better mentor for my students and gave me a sense of satisfaction. It’s also an extremely transferable skill: once you get good enough to use coding as a tool, it’ll become useful for many tasks that you hadn’t previously considered. I suggest you stick with it.
By the way, another advantage of working computationally is that no safety gear is required.